Download Center
Loupe Browser 8.0.0 (Mar 25, 2024)
- Download for WindowsFile size: 715 MBmd5sum: 4fc904fb79310e32333c1937a7b89205
- Download for MacOSFile size: 802 MBmd5sum: d629e9a085b3a49900e57052e8fa0cca
Download is unavailable on mobile devices.
Loupe Browser 8.0.0 highlights
- Enter the era of whole transcriptome analysis at high definition: Loupe Browser 8 supports manual alignment, visualization and analysis of Visium HD!
- Explore the relationships between cell types or pathways with the new co-expression functionality.
- Analyzing a large dataset? No problem. Loupe Browser 8 enables differential expression and reclustering analysis for unlimited barcodes or bins.
LoupeR package
10x Genomics' LoupeR is an R package that works with Seurat objects to create a Loupe Browser file. The Loupe Browser file can then be imported into Loupe Browser (v7.0 or later) for data visualization and further exploration.
Learn more about LoupeR packageAvailable for download on the 10x GitHub page
Tutorials
Explore the Loupe Browser tutorials for inspecting and investigating your Chromium Single Cell and Visium Spatial Gene Expression data.
System Requirements
Windows
- Windows 10 (64-bit) or later
- 16GB RAM
- SSD storage highly recommended
- Updated video/display drivers recommended
- 100% scaling display setting recommended
macOS
- macOS 10.15 (Catalina) or later
- 16GB RAM
- SSD storage highly recommended
- Default display settings recommended
Performance Notes
- When analyzing large single cell datasets (100k+ cells), 16GB RAM and a quad core processor are highly recommended for optimal performance.
- Loupe Browser v6.0 and later is compatible with .cloupe files produced by Cell Ranger v1.3+ and can import .vloupe files (VDJ clonotypes) produced by Cell Ranger v2.0+
Installation
Windows
Loupe Browser for Windows is distributed as a self-installing .exe file. Double-click on the downloaded file to install.
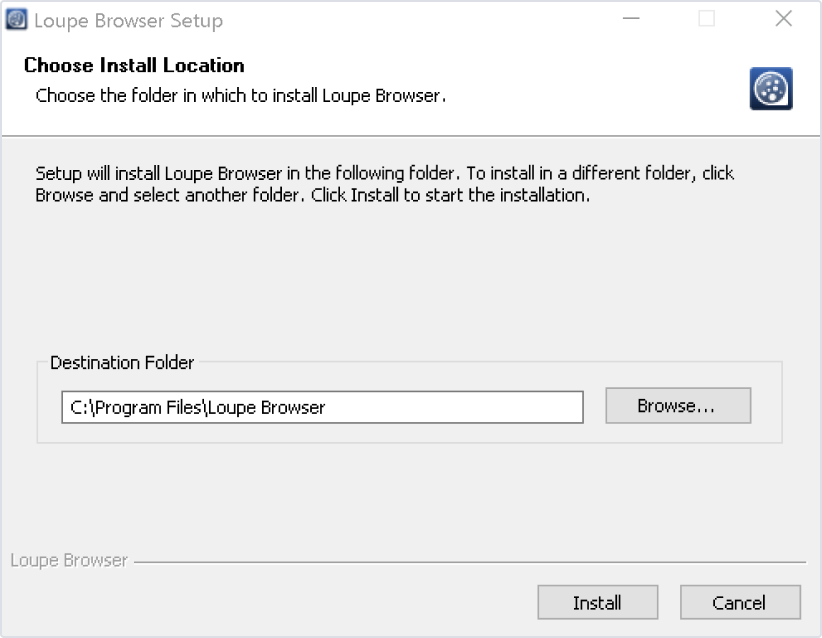
Follow the prompt to choose an installation folder. After installation, open Loupe Browser by double-clicking on the desktop icon, or double-clicking on a .cloupe file on your computer.
macOS
Loupe Browser for Mac is distributed as a .dmg file. Double-click to open the file. Then install Loupe by dragging the Loupe icon into the Applications folder. Open Loupe Browser by double-clicking on the app icon.
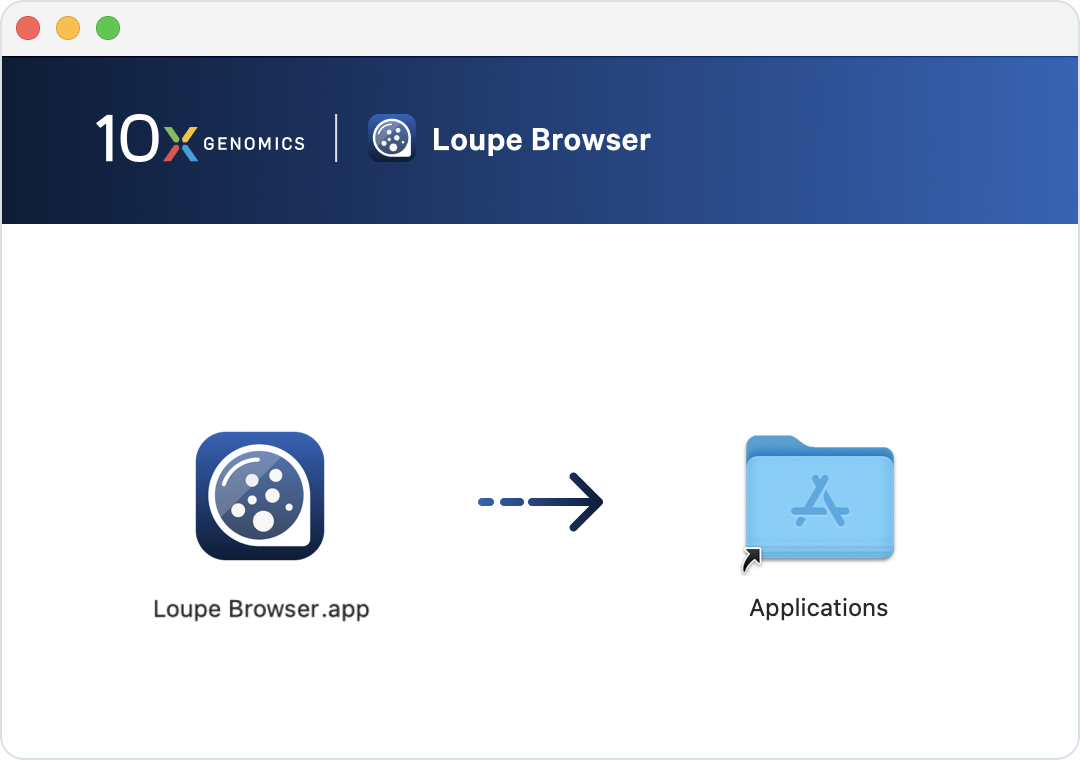
Loupe Browser can then be opened within the Applications folder or by double-clicking the Loupe Browser icon on the desktop.