Dissecting the composition of non-malignant cells within a tumor is key to understanding the interaction between a tumor and its microenvironment, predicting the clinical outcome, and assisting in the selection of therapies. In the Application Note, Characterization of the Tumor Microenvironment, we showed how the Single Cell Immune Profiling Solution was used to characterize a colorectal cancer (CRC) tumor and a squamous cell non-small cell lung carcinoma (NSCLC).
Here, we show how the NSCLC data was analyzed using Loupe Browser and Loupe V(D)J Browsers using the outputs generated by Cell Ranger. In this tutorial, we show how to:
- Identify and classify populations of cells based on gene expression profiles.
- Examine the repertoire of the immune cells.
- Link the gene expression with the immune receptor sequence for the same single cells.
- Generate the figures in the application note.
For this tutorial, and for the application note the data were analyzed using Cell Ranger v2.1, and visualization was performed using Loupe Browser v2.0.0 and Loupe V(D)J Browser v2.0.0.
In order to follow this tutorial you will need to download and install Loupe Browser and Loupe V(D)J Browser.
You also need some files from the NSCLC tumor data set (download links below). This data has already been run through Cell Ranger, a set of analysis pipelines that process Chromium single cell RNA seq and V(D)J reads. The RNA seq pipeline aligns reads, generates gene-cell matrices, and performs clustering and gene expression analysis. The V(D)J pipeline assembles the reads into TCR/Ig chains, annotates them, and generates clonotypes.
There are three data sets associated with the NSCLC tumor: 5’ gene expression, Ig enrichment from amplified cDNA (B cell Immunoglobulin (Ig) repertoire sequencing data) and TCR enrichment from amplified cDNA (T Cell Receptor (TCR) repertoire sequencing data). For each data set you need the Summary HTML to review the quality metrics and the Loupe file to open the file in the Browser.
Note that the .vloupe file you need to download will depend on the versions of Loupe Browser and Loupe V(D)J Browser you have installed. The first file in each section below is compatible with Loupe V(D)J Browser v4.0 and above, as well as Loupe Browser v5.0 and above. The second file should be used if you have Loupe V(D)J Browser v3.0 or below and Loupe Browser v4.2 or below.
These files can be downloaded using the following links:
5’ gene expression
Ig enrichment from amplified cDNA
TCR enrichment from amplified cDNA
In Table 1 of the application note (below) we have selected some of the key metrics that Cell Ranger outputs in the Summary HTML file to present a basic overview of the data quality.
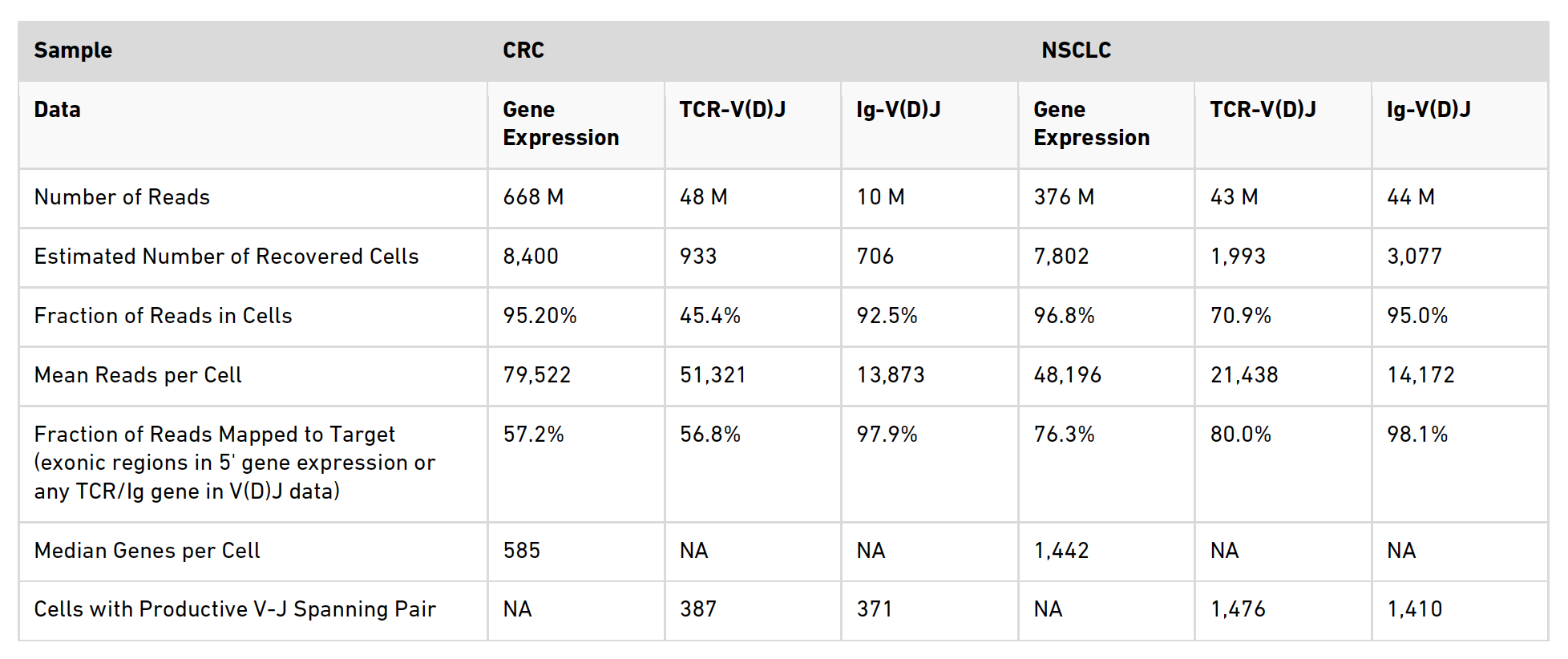
A summary of the metrics examined in the table is provided below.
Number of reads
This is the total sequencing reads for a sample.
Estimated Number of Recovered Cells
For the gene expression data this number is total number of barcodes associated with cell-containing partitions. Typically this is estimated from the barcode UMI count distribution. However, there is an assumption built into the cell calling algorithm that the RNA content between cells in a sample will only differ by an order of magnitude (based on empirical observations). When samples contain a highly heterogeneous mix of cells, as is the case in a tumor environment, this assumption is not correct and consequently Cell Ranger will under count the cells. To circumvent this you can specify the number of cells you want Cell Ranger to call using the --force-cells option. The number chosen to force cells to should be based on the expected cell number. However, defining the exact number can be an iterative process and you may have to try several different numbers of cells before getting the appropriate output. This is what we did in both samples here, hence the reported numbers are those that were used to force cells. For more information on the --force-cells option, see the Cell Ranger Algorithms Overview page. For the TCR and Ig data sets the expected number of cells is based on the number of cells with detected immune receptor transcripts and therefore, is expected to be lower than in the gene expression numbers as T cells and B cells only contribute to a fraction of the total cell number.
Fraction of Reads in Cells
Is the proportion of reads that are coming from cell barcodes. Reads that are not associated with a cell may be attributed to ambient RNA present in the sample. You can see that for the gene expression and Ig data sets for both samples that this metric is very high but it is lower for the TCR data sets, suggesting some background is present in this library. For more information on this metric see the FAQ page How to interpret 'Fraction of Reads in Cells' metric.
Mean Reads per Cell
This is the total number of sequenced reads divided by the estimated number of cells. Here, we targeted ~50,000 reads per cell in the gene expression data sets, however, depending on your cell type and application it is possible to use fewer reads than this. For more information on the number of reads per cell to use for gene expression data, see our Technical Note. For the repertoire profiling (both the TCR and Ig), we targeted ~5,000 reads per cell. You can see that we have exceeded this in some instances here, but this is not necessary.
Fraction of Reads Mapped to Target
This is the fraction of reads that map to exons for gene expression data sets (Reads Mapped Confidently to Exonic Regions), or to TCR/Ig sequences for the repertoire sequencing data (Reads Mapped to Any V(D)J Gene). Here, it is good for the NSCLC sample at around 76%, but is lower than expected in the CRC gene expression data. This is likely due to the high proportion of dying cells in this sample, see section 4.3 Identification of dying cells for further information on how these cells were classified.
Median Genes per Cell
This is the median number of genes detected (with nonzero UMI counts) across all cell-associated barcodes. This metric can provide an insight into the average transcriptional activity of the cells in the sample.
Cells with Productive V-J Spanning Pair
This is the number of cell barcodes for which at least 1 sequence was found for each of TRA and TRB in the case of T cells and the number of cell barcodes for which at least 1 sequence was found for each of IGH and IGL in the case of B cells.
Loading Data and Clustering Cells
To visualize the gene expression data, open Loupe Browser and import the Loupe Browser file that you have downloaded. To open the file go to: File > Open File. Alternatively, you can double click on the downloaded .cloupe file to open it.
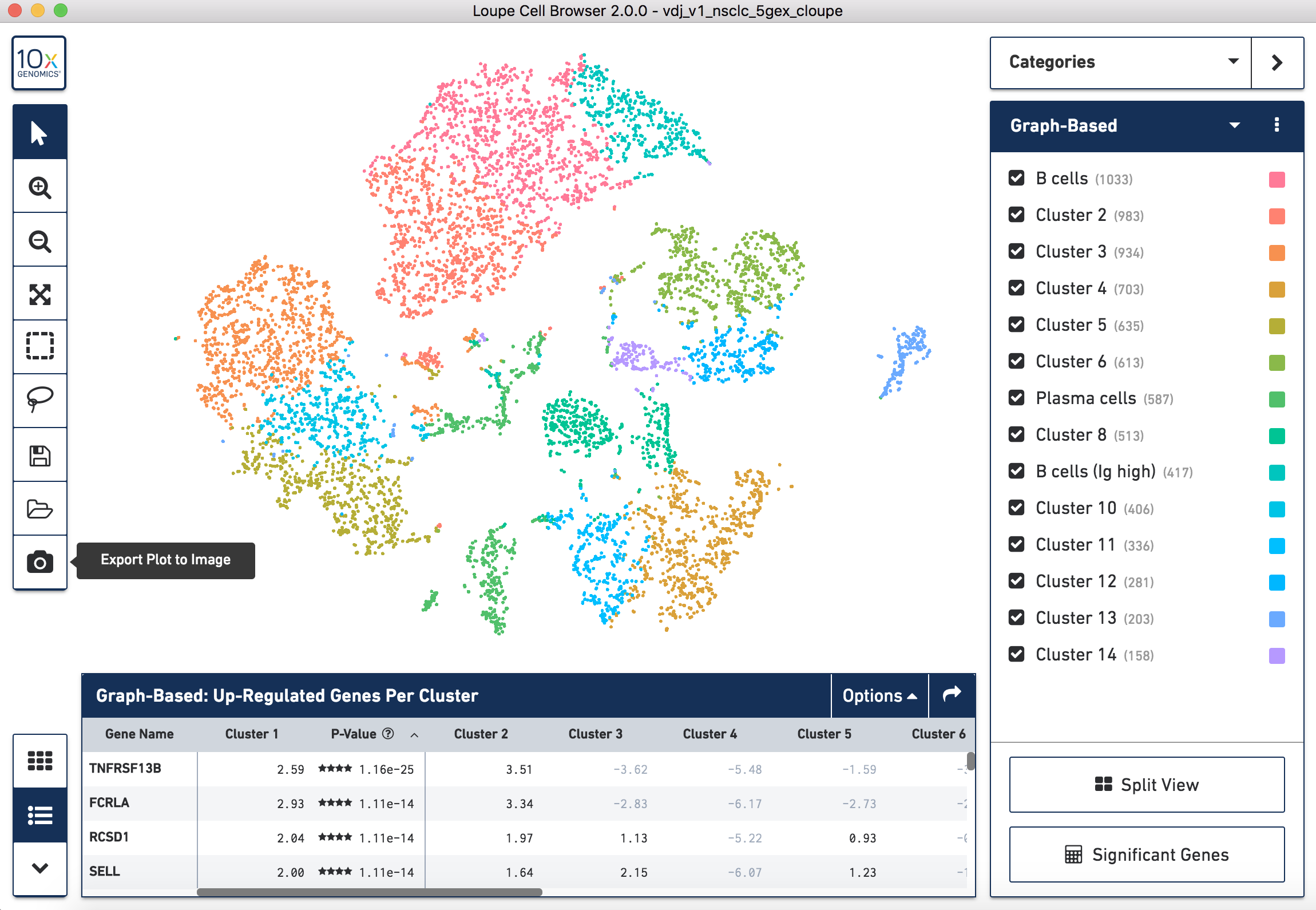
When you open the file, Loupe Browser will display the t-SNE plot generated by Cell Ranger. Each cell is represented by a dot and cells are positioned such that they are close to other cells with similar gene expression patterns. In the application note, we use the default Graph-Based clustering algorithm to identify the clusters within the t-SNE plot, but this can be changed to K-Means clustering by toggling the selection in the upper right menu bar.
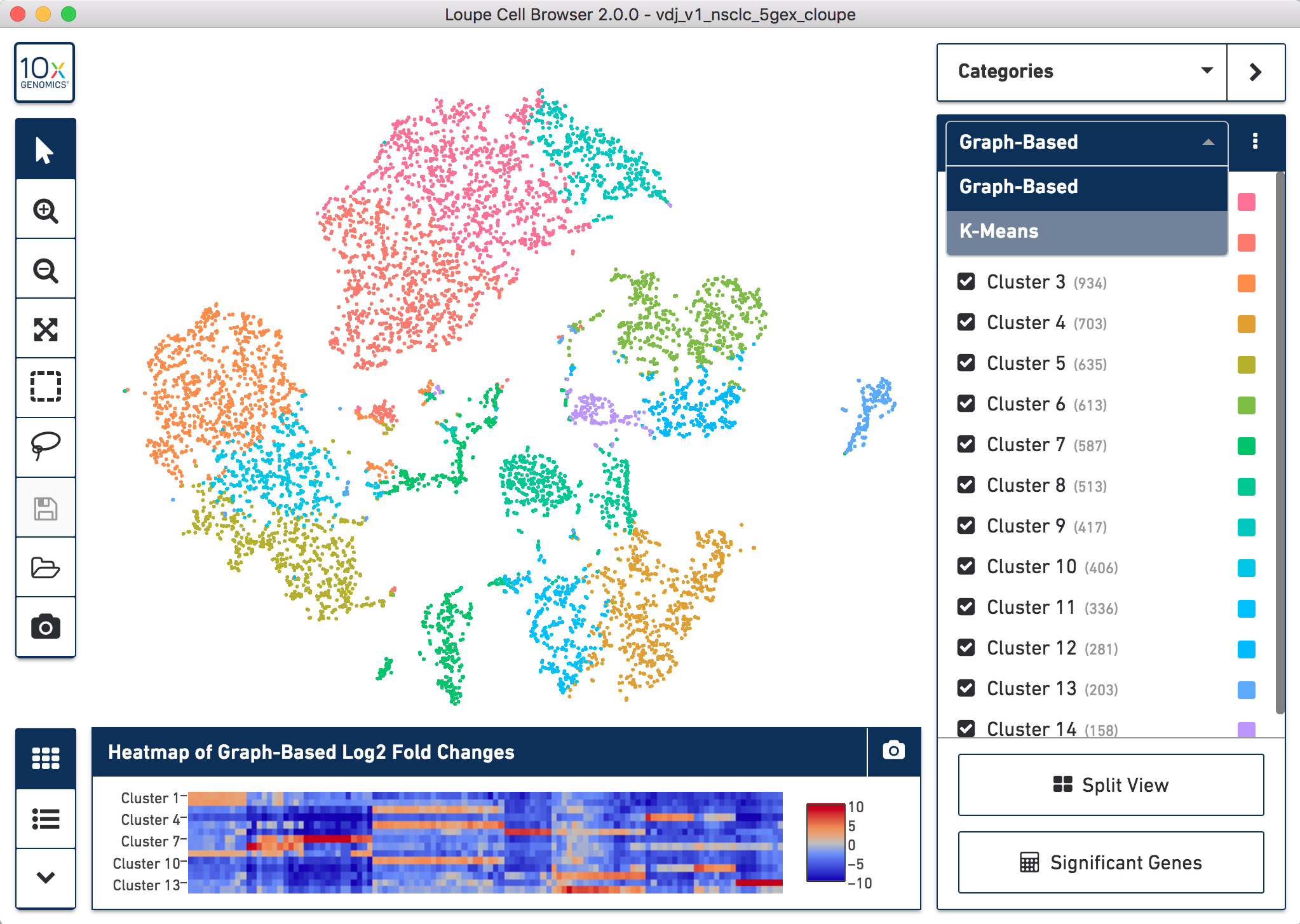
To perform cell type classification for the different clusters, we looked at the gene expression profiles of the cells. This can be done by examining the listed upregulated genes for a specific cluster and/or by looking at the expression of known gene markers for a particular cell population. Clusters are then manually classified based on this information. Assigning cell types to clusters is an iterative process that requires looking at the gene expression patterns and coming to conclusions about the dominant cell type driving the gene expression in a particular cluster.
We show you how to do this using Loupe Browser in the video below:
Note: There will be genes you have not heard of in the gene lists for each cluster; look them up and find out what they do! Some useful websites for exploring gene functions are listed in section 7.1 Useful websites for exploring gene functions.
To perform cell type classification for the different clusters, we looked at the gene expression profiles of the cells. This can be done by examining the listed upregulated genes for a specific cluster and/or by looking at the expression of known gene markers for a particular cell population. Clusters are then manually classified based on this information. Assigning cell types to clusters is an iterative process that requires looking at the gene expression patterns and coming to conclusions about the dominant cell type driving the gene expression in a particular cluster.
We show you how to do this using Loupe Browser in the video below:
Note: There will be genes you have not heard of in the gene lists for each cluster; look them up and find out what they do! Some useful websites for exploring gene functions are listed in section 7.1 Useful websites for exploring gene functions.
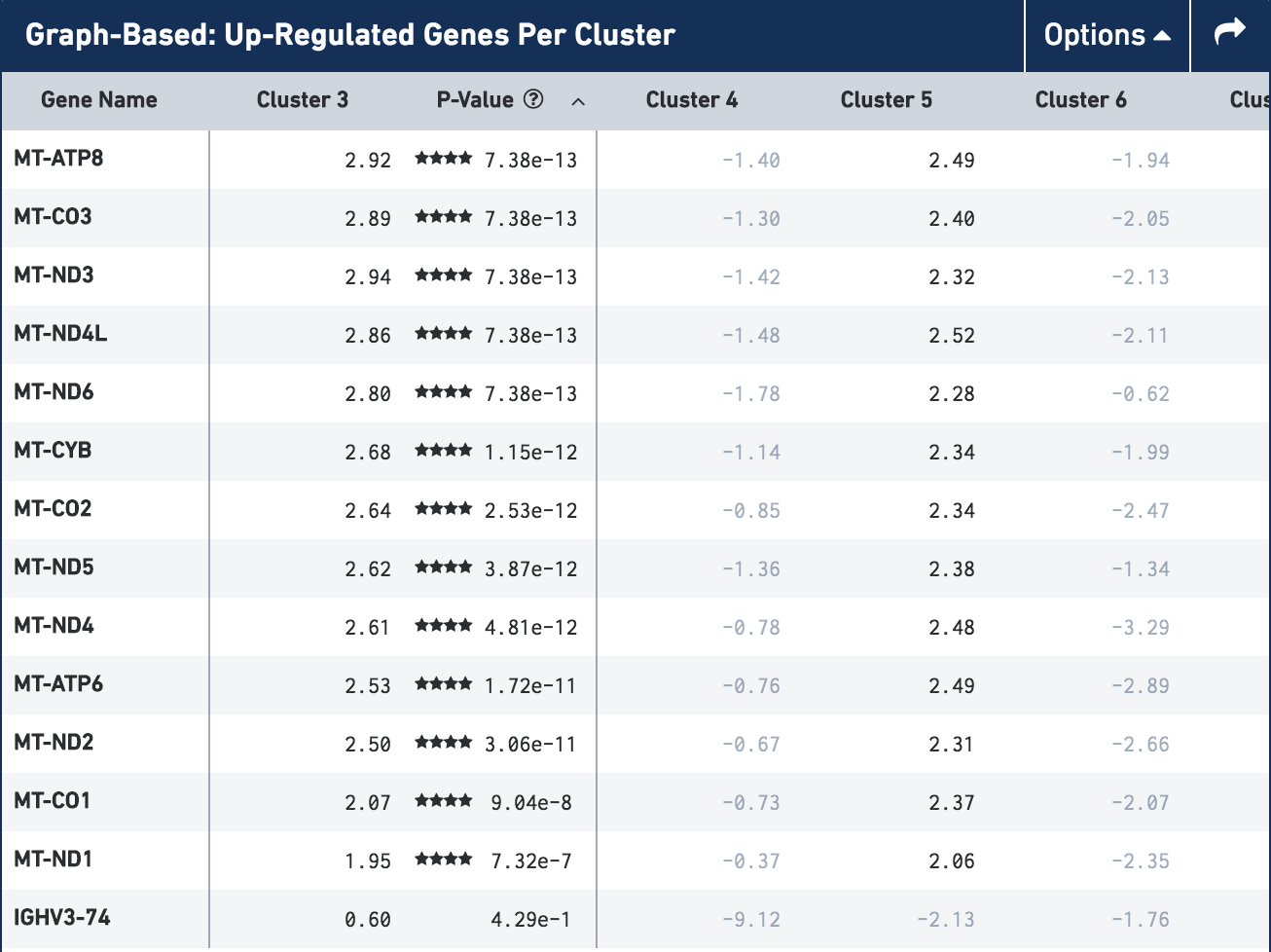
A common question that arises when performing cell type classification, is how do we identify dead or dying cells? In the CRC sample, we identified three clusters as dying cells (Figure 1 of the application note). When we look at the gene expression profiles of those clusters there are two things that stand out:
- There are very few genes that are upregulated (indicating low overall gene expression);
- Those that are upregulated are mitochondrial genes (prefixed with MT).
These two factors are indicative of poor cell health, suggesting that the cells are either about to enter or are already going through apoptosis or necrosis pathways.
In the image below you can see the gene table for cluster 3 of Graph-Based clustering in the CRC t-SNE plot (the orange cluster in the top right). In line with the criteria for identifying dead cells above, there are only 14 genes that are upregulated in this cluster and all of those that are significantly upregulated are mitochondrial genes. The expression of the genes that are upregulated in cluster 3 can also be seen in cluster 5 (the yellow/green cluster in the center), another cluster we classified as 'Dying cells'.
To generate your figure you can export a .png of the t-SNE plot, click the Export Plot to Image camera icon at the bottom of the left hand toolbar. Once you have exported the image, you will need to add labels manually with the cell type classification you have decided on.
The Single Cell Immune Profiling Solution also allows you to profile the clonotypes of the T cell receptor (TCR) and B cell immunoglobulins (Ig) from the same sample. We can examine these clonotypes in detail using Loupe V(D)J Browser. Here, we will use the corresponding TCR and Ig clonotype data from the NSCLC sample that we downloaded.
To visualize the data we can import the TCR or Ig Loupe V(D)J Browser file into Loupe V(D)J Browser. To open the file go to File > Open File. Alternatively, you can double click on the downloaded .vloupe file to open it.
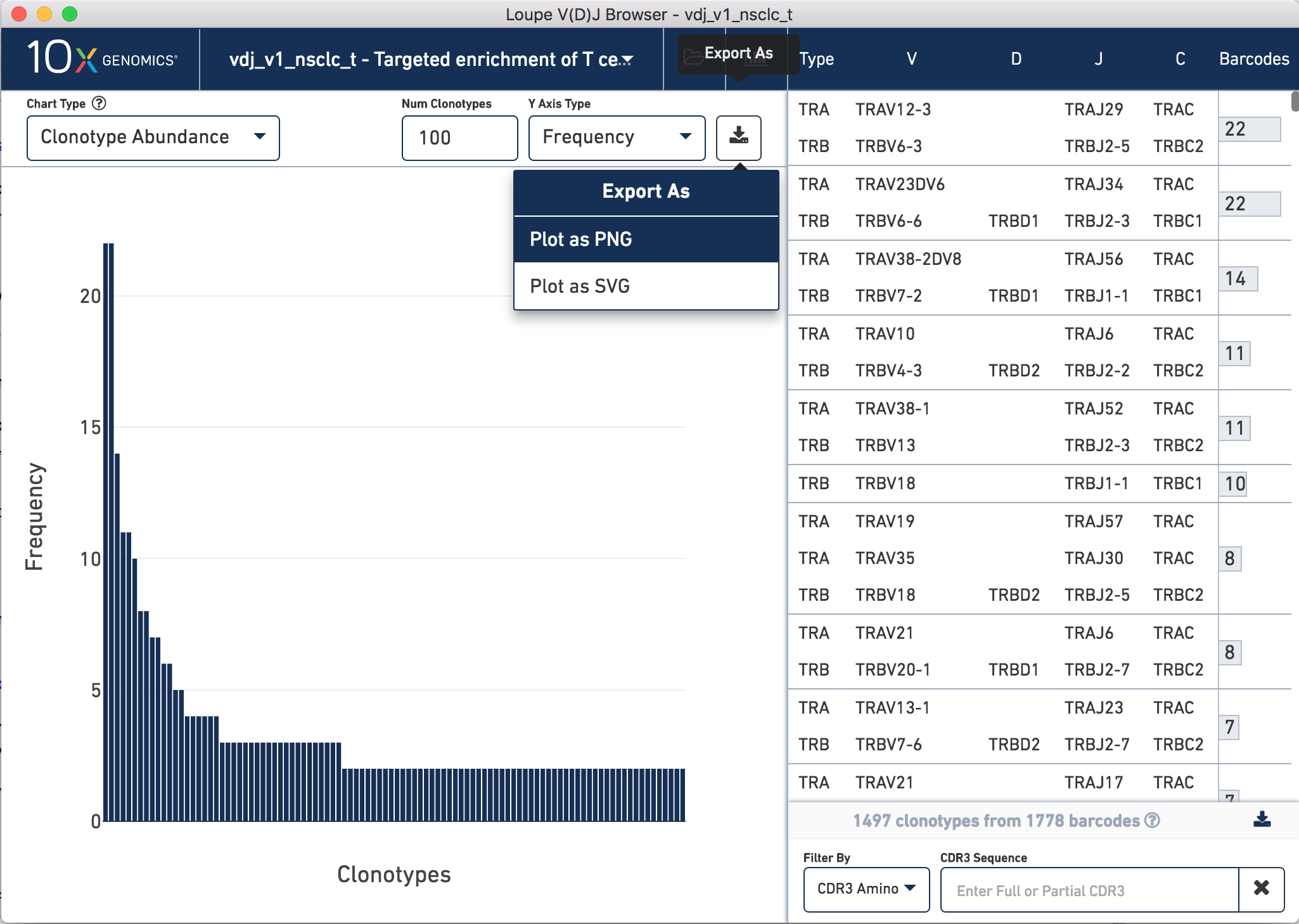
The landing view is a histogram of the frequency distribution of different clonotypes. These plots were used in the application note to generate Figure 2.
Looking at the clonotype distribution is only a very small part of the functionality of Loupe V(D)J Browser. For a comprehensive tutorial on how to explore your repertoire sequencing data, check out the Loupe V(D)J Browser tutorial.
To generate Figure 2, we exported the clonotype distribution plots for both TCR and Ig data sets for our NSCLC and CRC samples. To do this, open your file of interest, click the Export As button in the top right hand corner of the plot, click Plot as PNG.
One of the most powerful things about the Single Cell Immune Profiling Solution is the ability to intersect the gene expression and the repertoire sequencing data, allowing you to not only understand the cell type based on the cluster classification, but also the associated clonotype for a specific cell.
This video shows how to do this in Loupe Browser:
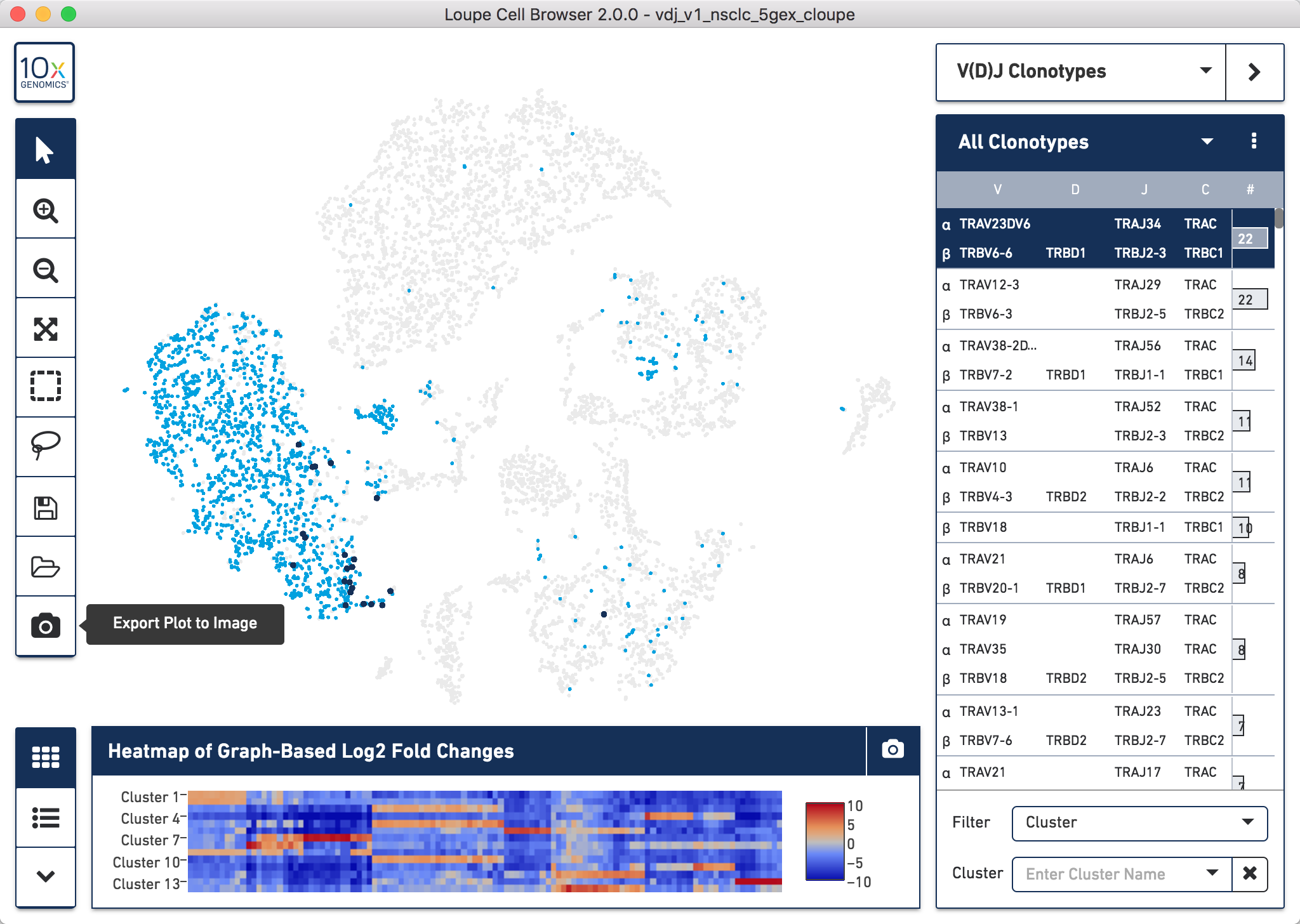
To generate Figure 3, we exported the image of the Ig clonotypes (light blue dots) with the dominant clone highlighted (dark blue dots), overlapped onto the t-SNE plot for the CRC sample. We then manually overlayed the annotation for the dominant clonotype onto the t-SNE plot.
In the example below we did this for the TCR clonotypes for the NSCLC sample, but the process is the same irrelevant of sample used. Once you have loaded the data as per the video in section 6.1, you can easily do this by clicking the Export Plot to Image camera icon at the bottom of the left hand toolbar.