Loupe Browser is a desktop application that provides programming free interactive visualization functionality to analyze data from different 10x Genomics solutions. Loupe Browser allows you to easily interrogate different views of your 10x Genomics data to quickly gain insights into the underlying biology. Loupe is named for a jeweler's loupe, which is used to magnify and inspect the details of precious gemstones. Loupe Browser supports the analysis of data from the 3' and 5' Chromium Next GEM Single Cell Gene Expression, Feature Barcode, Single Cell ATAC, Single Cell Multiome ATAC + Gene Expression, and Visium Spatial Gene Expression solutions.
Loupe Browser provides many powerful analysis capabilities. Several tutorials have been provided to walk you through its features and uses for analyzing 3' and 5' Gene Expression and Feature Barcode data. Start your journey with the Single Cell Navigation tutorial.
The Loupe Browser ATAC tutorials demonstrates how to find significant features, analyze differential accessibility patterns, and explore substructure within a real-world dataset.
The Loupe Browser Multiome ATAC and Gene Expression tutorials review the features and functionality unique to data generated by Cell Ranger ARC and the Multiome ATAC + Gene Expression kit, and introduce techniques and patterns for analyzing linked gene expression and open chromatin regions.
The typical spatial data analysis workflow involves Loupe Browser at both upstream and downstream steps. In addition to the downstream visualization and analysis capabilities, Loupe Browser offers support for manual alignment of fiducial frame and tissue selection upstream of running Space Ranger pipeline.

To use Loupe Browser, open any .cloupe file generated by the Space Ranger analysis
software which is embedded with the following information:
- Space Ranger generates a tiled version of the tissue image to enable fast zooming.
- Gene expression information for all spots on the slide.
- Information about spot alignment in the image.
- Various gene expression-based clustering information for the spots, including t-SNE and UMAP projections.
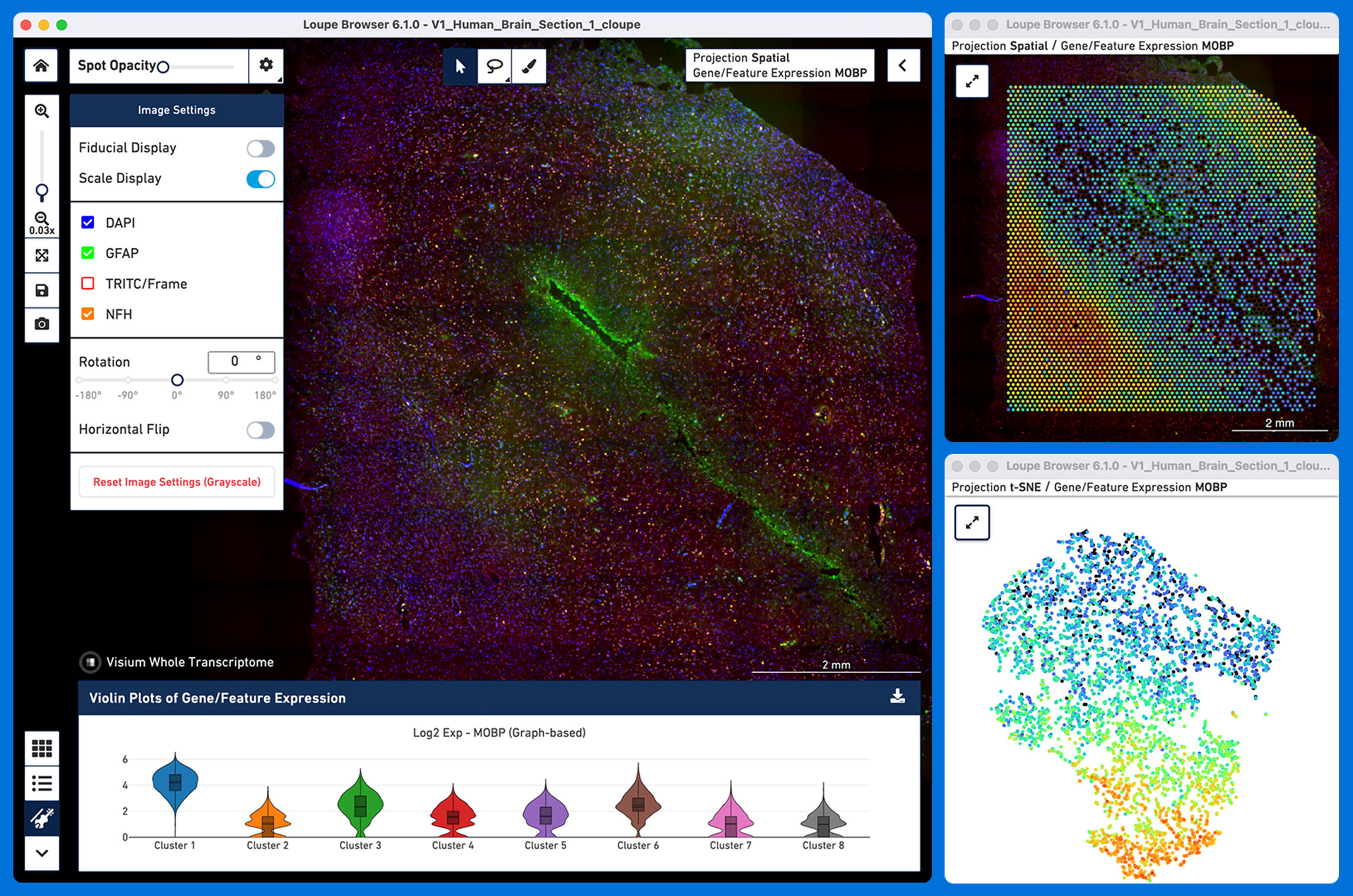
Loupe Browser provides many powerful analysis capabilities including the ability to perform the following analysis all within the spatial context of a tissue image:
- Interrogate significant genes
- Characterize and refine clusters
- Perform differential expression analysis
To learn how to use Loupe Browser to analyze your gene expression data, review the following tutorials:
-
Manual Fiducial Alignment: Use Loupe Browser guided fiducial alignment and tissue selection tools (version 4.0 and later) to generate the manual alignment file in JSON format necessary for
spaceranger countpipeline. -
Manual CytAssist Image Alignment: Use the Loupe Browser-guided landmark selection and auto refine tools (version 6.2 onwards) to align the CytAssist captured image on the Visium slide with a microscope image of the same tissue section on a standard glass slide to generate an alignment JSON file necessary for spaceranger count pipeline.
-
Navigation: Understand the key features of the interface, the different views, modes, and data selector panels as well as export and import options.
-
Evaluate Gene Markers: Use gene markers of known cell type to view their spatial distribution and explore the use of gene expression independent spatial enrichment metric Moran's I (version 5.1 and later) to evaluate spatiality.
-
Explore Spatial Clusters: Understand gene expression data in a spatial context and use gene lists to create custom categories and evaluate differential gene expression between them.
-
Characterize Substructure: Identify subpopulations within clusters in the dataset and create complex Boolean filters to locate regional subfields within a cluster.
-
Spatial Reclustering: Reanalyze spatial datasets for clusters or regions of interest by customizing clustering and projection parameters to identify new insights.
-
Sharing Results: Save features of interest, export data tables, and plots from the Visium Spatial data.
Starting with Loupe Browser v6.1, users may choose to share anonymized non-biological usage data (i.e. no identifiable sample information such as files, cluster names, or any open text fields are tracked) to improve Loupe Browser performance. Sharing of usage data is optional, and users can change whether they want to share usage data any time. The user preference for usage data sharing will carry over within a major version (e.g. all 6.Y versions) but the user will be asked to confirm their preferences for each major version update.
In case you wish to change the usage data sharing setting, click Manage Usage Data from the Help menu and change the preference in the pop-up window.