Detect chromatin accessibility
Analyze thousands of unique open chromatin fragments per cell, genome-wide.
Single cell resolution
Go beyond population averages by measuring epigenomic profiles in single nuclei.
Flexible and scalable
Profile hundreds to tens of thousands of nuclei per chip.
Streamlined data analysis
Explore and visualize chromatin accessibility profiles with easy-to-use software.
One-day lab workflow
From sample to sequencing-ready library in one day.
Diverse sample compatibility
Demonstrated with cell lines, primary cells, and fresh and frozen tissue samples.
Explore what you can do
Define cell types and states
Characterize regulatory networks
Discover cis-regulatory elements, such as promoters and enhancers
Reveal associations with treatment response
Proven Results
Publications
Developmental and evolutionary dynamics of cis-regulatory elements in mouse cerebellar cells
Developmental and evolutionary dynamics of cis-regulatory elements in mouse cerebellar cells
Science. 2021, Sarropoulos I, et al.
Science. 2021, Sarropoulos I, et al.
Cell-type specialization is encoded by specific chromatin topologies
Cell-type specialization is encoded by specific chromatin topologies
Nature. 2021, Winick-Ng W, et al.
Nature. 2021, Winick-Ng W, et al.
Single-cell analyses reveal key immune cell subsets associated with response to PD-L1 blockade in triple-negative breast cancer
Single-cell analyses reveal key immune cell subsets associated with response to PD-L1 blockade in triple-negative breast cancer
Cancer Cell. 2021, Zhang Y, et al.
Cancer Cell. 2021, Zhang Y, et al.
Resources
Find the latest app notes and other documentation Chromium Single Cell ATAC.
Is single cell epigenomics right for me? ATAC-ing your research questions for deeper insights
Is single cell epigenomics right for me? ATAC-ing your research questions for deeper insights
10x Genomics Blog
10x Genomics Blog
Deciphering Epigenetic Regulation with Single Cell ATAC-Seq
Deciphering Epigenetic Regulation with Single Cell ATAC-Seq
Application Note, 10x Genomics
Application Note, 10x Genomics
Single Cell ATAC-Seq for Characterization of Complex Biological Systems
Single Cell ATAC-Seq for Characterization of Complex Biological Systems
Application Note, 10x Genomics
Application Note, 10x Genomics
Our End-to-End Solution

Chromium Single Cell ATAC Reagents
With our reagent kits, detect open chromatin regions using the included transposase, and generate ready-to-sequence NGS libraries.
Analysis and Visualization Software
Our analysis pipelines
Our visualization software

World-Class Technical and Customer Support
Our expert support team can be contacted by phone or email at support@10xgenomics.com
Workflow
- 1
Prepare your sample
Start with a nuclei suspension isolated from cell culture, primary cells, or fresh or frozen tissue.
CHROMIUM NUCLEI ISOLATION KIT
For a wide range of frozen mammalian tissues, the Chromium Nuclei Isolation Kit offers an all-in-one kit for easy and reliable nuclei isolation optimized for the 10x Genomics Single Cell ATAC assay.
Resources - 2
Construct Your 10x Library
Construct a 10x barcoded library using our reagent kits and a compatible Chromium instrument. Each member of the Chromium instrument family encapsulates each cell with a 10x barcoded Gel Bead in a single partition. Within each nanoliter-scale partition, cells undergo reverse transcription to generate cDNA, which shares a 10x Barcode with all cDNA from its individual cell of origin.
Resources - 3
Sequence
The resulting 10x Barcoded single cell ATAC-seq library is compatible with standard NGS short-read sequencing on Illumina sequencers, for massively parallel epigenomic profiling of thousands of individual cells.
Resources - 4
Analyze Your Data
Our Cell Ranger ATAC analysis software generates open chromatin profiles for each cell, identifies clusters of cells with similar profiles and calls peaks, and can aggregate data from multiple samples.
Analysis Pipelines Output
Output includes QC information and files that can be easily used for further analysis in our Loupe Browser visualization software, or third-party R or Python tools.
Resources - 5
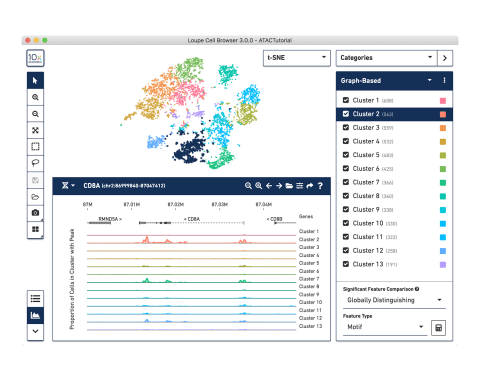
Visualize Your Data
Use our Loupe visualization software to interactively explore your results, perform differential accessibility analysis, find clusters enriched for motifs of interest, and compare across samples.
Do I need to be a bioinformatician to use it?
Loupe is a point-and-click software that’s easy for anyone to download and use.
Resources
Frequently Asked Questions
It enables profiling of the open chromatin landscape at single cell resolution.
Yes, using third party tools. For more information, watch our webinar.
A nuclei suspension is needed. It is possible to obtain high-quality nuclei suspensions from fresh and cryopreserved cells, fresh tissue, and frozen tissue. The Chromium Nuclei Isolation Kit can be used to generate high-quality nuclei suspensions from a wide range of frozen tissues with minimal optimization.
This assay has been optimized for human and mouse samples, although other species may be possible.
Single cell ATAC-seq enables the study of highly heterogeneous samples, identifying unique subpopulations of cell types based on their open chromatin profiles. This could include groups of cells at different developmental stages. Bulk ATAC-seq can only provide an average readout of open chromatin from your sample, potentially masking this cellular heterogeneity.
ATAC-seq captures all open chromatin regions, not just those bound by a specific factor. It is an unbiased (genome-wide) approach to look for epigenetic changes in a sample. For proteins with a known sequence binding motif, the included software can identify those motifs that are enriched in open chromatin on a cell-by-cell basis.