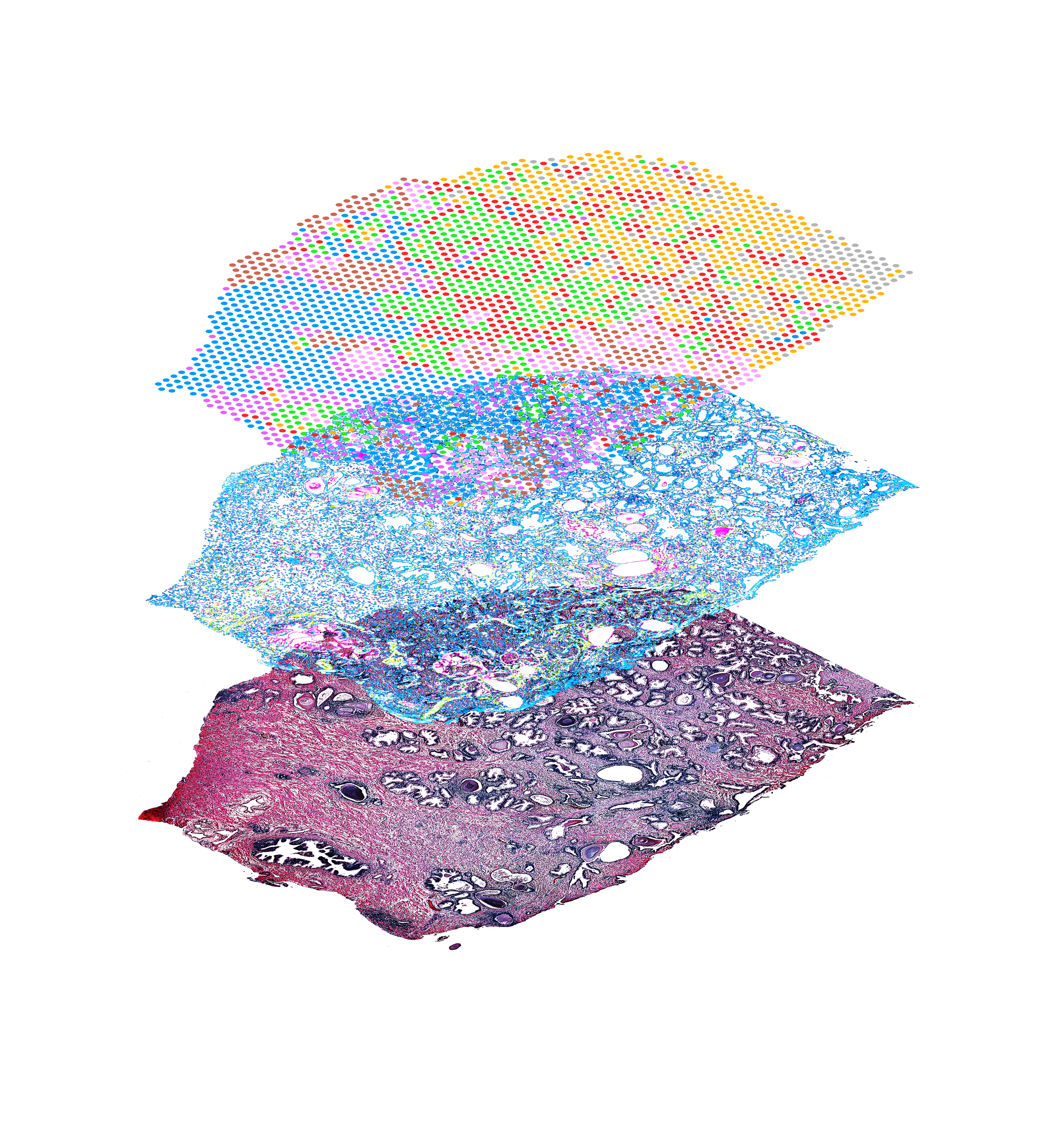
Maximize your insights
Gain a deeper, more complete assessment of your samples with protein, mRNA, and morphology from the same tissue section.
Profile entire tissue sections
Visualize protein detection and whole transcriptome expression, without the need to select regions of interest.
Streamlined data analysis
Combine protein detection profiles with gene expression data in easy-to-use software.
Explore what you can do
Gain a complete view of disease complexity
Discover new biomarkers and identify novel cell types and states
Map the spatial organization of cell atlases
Identify spatiotemporal gene expression patterns
Rich insights
- Single-section multiomics - Study both protein and RNA from a single tissue section using a curated, pre-validated antibody content panel
- Histology matters - The only multiplex protein assay that allows you to image an H&E or IF-stain before other chemical treatments
- Bias-free discovery - Unbiased gene expression analysis with whole transcriptome detection and no need for ROI selection with predetermined biomarkers

Proven results
Publications
Transcriptome-scale spatial gene expression in the human dorsolateral prefrontal cortex
Transcriptome-scale spatial gene expression in the human dorsolateral prefrontal cortex
Maynard KR, et al. Nature (2021).
Maynard KR, et al. Nature (2021).
Mapping the developing human immune system across organs
Mapping the developing human immune system across organs
Suo C, et al. Science (2022).
Suo C, et al. Science (2022).
Single-cell and spatial analysis reveal interaction of FAP+ fibroblasts and SPP1+ macrophages in colorectal cancer
Single-cell and spatial analysis reveal interaction of FAP+ fibroblasts and SPP1+ macrophages in colorectal cancer
Qi J, et al. Nature Communications (2022).
Qi J, et al. Nature Communications (2022).
Resources
Find the latest app notes and other Spatial Gene and Protein Expression documentation
The complete toolkit

Visium CytAssist
A compact instrument to facilitate the transfer of transcriptomic probes from sectioned tissues to Visium Slides.
Visium CytAssist Gene and Protein Expression reagents
With our reagent kits, capture whole transcriptome gene and protein expression with spatial resolution across an entire tissue section.
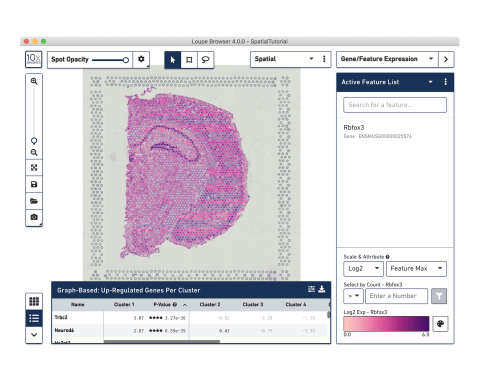
Analysis and visualization software
Our analysis pipelines
Our visualization software

World-class technical and customer support
Our expert support team can be contacted by phone or email at support@10xgenomics.com
Frequently asked questions
The Visium CytAssist Spatial Gene and Protein Expression assay is designed to introduce simultaneous Gene Expression and Protein Expression analysis to FFPE samples processed with Visium CytAssist. The assay uses NGS to measure the abundance of oligo-tagged antibodies with spatial resolution, in addition to the whole transcriptome and a morphological image.
Our Visium CytAssist Gene and Protein Expression solution allows for researchers to study the relative expression patterns of proteins within the Human Immune Cell Profiling Panel. An NGS read-out of Protein Expression data allows you to overcome challenges like spectral overlap and restricted antibody host species, which generally lead to limitations in the number of proteins that can be studied simultaneously in tissue. Furthermore, it enables users to interrogate immune protein markers that, typically, are lowly expressed as RNA.
Our panel features 35 antibodies from Abcam and Biolegend, and includes cell surface and intracellular targets. For a list of antibodies included, please see the Antibody List on the Support site or in the appendix of this FAQ.
10x Genomics provides two types of software to help you analyze your data: Space Ranger and Loupe Browser. Space Ranger is an analysis software that automatically overlays spatial gene expression information on your tissue image and identifies clusters of spots with similar transcription profiles. You can then use Loupe Browser, a visualization software, to interactively explore the results. You can download Space Ranger and Loupe Browser from the 10x Genomics Support site at no cost.
Yes, there are several key differences between standard IF and the IF protocol used with Visium. Please refer to the IF Demonstrated Protocol (FFPE | Fresh frozen) for additional details.
Impact on Visium performance may vary from tissue to tissue due to the high sensitivity of the Visium assay. However, the spatial organization of gene expression and cell cluster detection is comparable between H&E and IF.